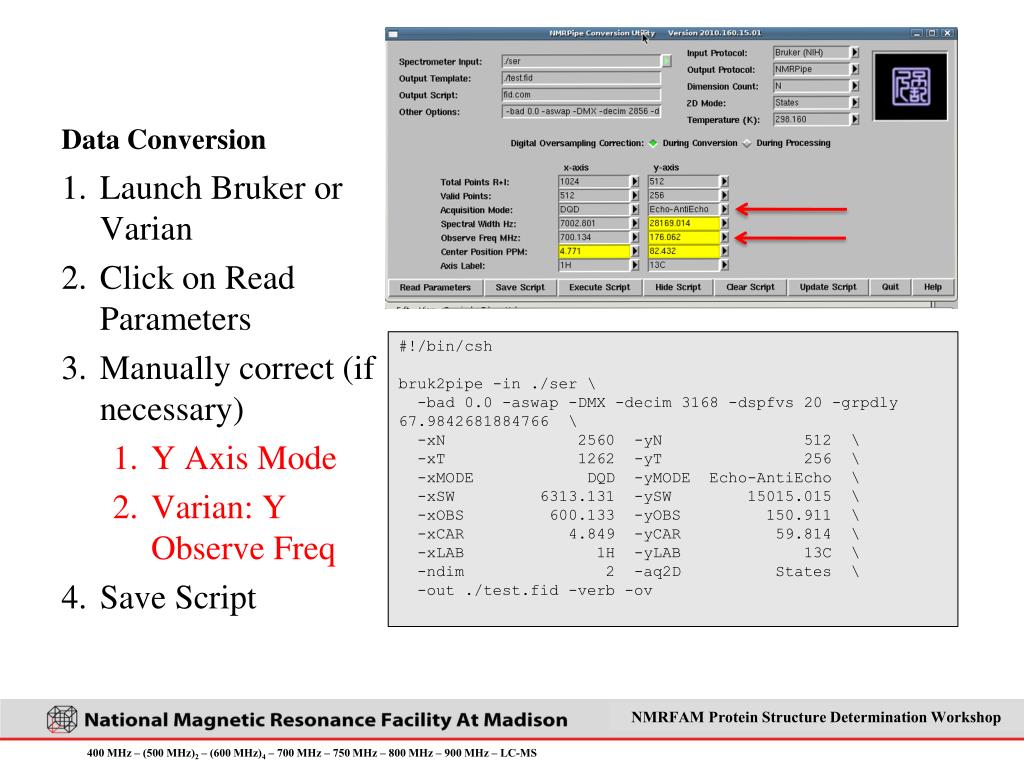
Save data in nmrpipe format using the extension “.fid” 2. Using the Delta File Browser select the above file and open it. Delta will automatically execute conversion from nmrpipe to Delta format ★ The.fid file should be a single file. If nmrpipe data are in the form of a series of files then combine them into a single file. The NMRPipe system is a UNIX software environment of processing, graphics, and analysis tools designed to meet current routine and research-oriented multidimensional processing requirements, and to anticipate and accommodate future demands and development.
2D NMR DataProcessing with nmrPipe
nmrPipe is a widely used, free, powerful NMR dataprocessing software. nmrDraw is the packaged graphic userinterface for data processing and display. nmrPipe is a Unix pipe command basedprogram for streaming FID and data vectors during processing of amulti-dimensional spectrum. The data pipes feed results from previous commandsto following commands, mostly in memory, to give the final processed data. Theprogram can process data from 1D to 4D. This document limits the discussion to2D data only.
Currently, the program runs under the Unix environment, including Linux, andunder Mac and Windows XP operating systems. The programs have been installed onNMR500, NMR600, and the data workstations. For more information on softwareavailability and references, visit author Frank Delaglio's website at NIH and his private website.
While learning the whole programming package takes time, I have written twoprograms (pipe2d or snap2d) thatautomatically generate scripts and do the processing for known types of routine2D experiments. You do have to adjust phase parameters for phase-sensitivespectrum after the initial processing. Please report problems you may encounterusing these programs.
The following details the procedure using pipe2d orsnap2d. For details of nmrPipe and nmrDraw program, see thefollowing pages.
See Section 3 for plotting with X and Y projections.
In the following, all commands must be entered in a Linux terminal window.To open a terminal, right-button click inside the Desktop, select OpenTerminal.
1: Procedure (after experiment finishes and datasaved)
See section below for taking snapshot during a 2D experiment run.
Bootcamp not enough space mojave. Says I do not have enough space for bootcamp. Posted by 1 year ago. Says I do not have enough space for bootcamp. Hello, I am a Mojave user. I was trying to install bootcamp and it said I needed at least 65GB of space to install. The Radeon drivers for Bootcamp were recent, but not recent enough for. Per Install Windows on your Mac with Boot Camp - Apple Support, I should have at least 64 GB available for the partition and I have that. One possible issue per If Boot Camp Assistant gives a 'Not enough space' error - Apple Support, flash install media that has been converted will have 32 and 64 bit images. That shouldn't be my issue because I have an ISO direct from MS. It is possible that there is not enough free space in a single congruent area on your SSD - meaning that fragmentation is keeping you from being able to partition. In that case, you can either try to find a utility that will defrag, or back up to another drive, reformat, partition, then restore. The Boot Camp Assistant will not install Windows because there really is not enough free space. One possible solution would be to use a 16 GB or larger flash drive to hold the Windows installation files. Once the files are transferred to the flash drive, you could delete them from the macOS volume and thus create more free space. When clicking continue on the first Boot Camp screen, I am presented with a dialog that displays: The startup disk does not have enough space to be partitioned. You must have at least 39 GB of free space available. This is despite 'About this Mac' reporting 271GB of free space: And 'Disk Utility' reporting 268GB of free space.
Reference the direct 1H dimension before saving data
Make sure that before you save your data, reference the 1H spectrum of thedirect dimension carefully in vnmrJ. The pipe2d program usesthe 1H reference of the direct dimension to reference the indirect dimension,regardless of the type of nucleus (1H, 13C, 15N, etc.) along the 2nddimension.
If you use the experiment setup procedure given in the instructions, thereference should be carried over from the 1D 1H spectrum done before the 2Dexperiment. No additional action is needed.
Run pipe2d
The pipe2d program must be launched from within a .fidfolder (we'll use noesy2d.fid as an example below) that containsVarian's data files. Enter the following commands:
- cd noesy2d.fid
- pipe2d
Enter the index number of the template experiment that matches the type ofexperiment you have. If you simply hit the RETURN/ENTER key, by default theprogram uses 0 (the 1st template experiment). The default value is correct mostof the times. Next, the program creates fid.com (dataconversion) and nmrproc.com (Fourier Transform) under the .fidfolder and executes the files to create the processed spectrum(test.ft2). Once done, the program automatically launchesnmrDraw. Press r with cursor in main window to draw contours.Both scripts can be executed manually, after manual editing of certainparameters, from the terminal window, by typing either fid.comor nmrproc.com. Data conversion usually only needs to be doneonce; processing may need to be re-run and fine tuned more times.
Adjust phases
There is no need for phase adjustment if the data is in
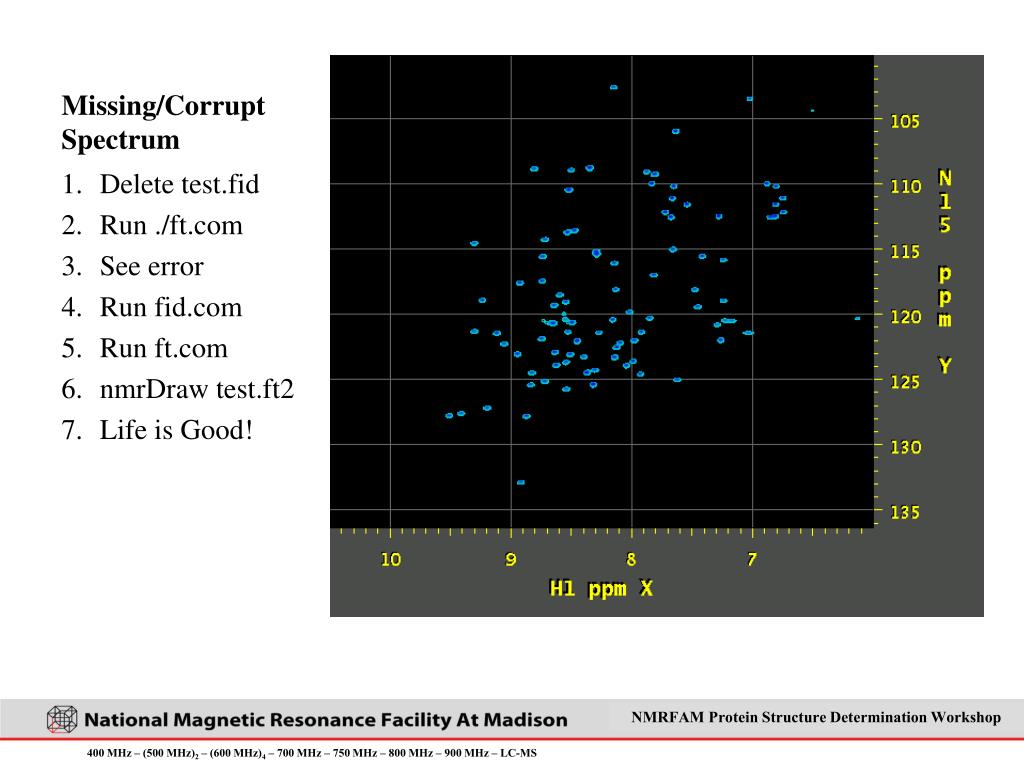
- pipe2d launches nmrDraw after data processing. If not, from within the .fid folder, type nmrDraw& to launch nmrDraw
- Put cursor inside the drawing area and press r to draw the contours. Click the +/- buttons next to Factor to adjust contour level to see all peaks. Press '-' (lower the contour threshold) and r to redraw contours until baseline noise starts to show. Then, press '+' to increase the contour level a bit so that baseline noise is barely visible and the positive/negative tails of peaks are visible. That's where you want to stay to make the rest of the adjustments.
- Check phase of the peaks along both X and Y axes. Out-of-phase peaks show uneven shoulders, bases, and positive/negative tails. If phases need correction, do the following:
- Right button click Mouse->1D Horizontal to bring out a horizontal line. Hold the left button to drag the line across peaks. To increase the vertical scale of the 1D slice, hold the middle button and click inside the grey area OUTSIDE the main graphic area (black), and drag the mouse up or down. To move the slice position in the window up or down, click and hold the right button and drag in the grey area.
- To make phase adjustment, toggle Phasing button to ON. Drag the P0 (zero order phase) bars (left bar: coarse; right bar: fine) until the base of the peak(s) is even and symmetric with the peak either pointing up or down.
- Click Mouse->1D Vertical to adjust phase for peaks along Y axis.
- If peaks across X or Y axis appear to need different amount of zero order phase correction, it means a linear (1st order) phase correction is required. The linear phase change shows as a linear increase or decrease of phase distortion along X or Y axis. Do the following:
- Bring out a horizontal or vertical slice. First, pick a slice that cuts a strong peak near the left edge of spectrum along X, or the bottom edge along Y. Set the Pivot point to this peak by clicking the Pivot +/- button. Make the yellow arrow point to the center of the peak.
- Adjust P0 so that this peak is phased perfectly.
- Then, move the slice to cut peaks at the other edge of the spectrum (right edge for X and top edge for Y axis). This should show out of phase peaks. Adjust P1 so that these peaks are phased nicely.
- Once P1 adjustment is done, select a slice across the center of the spectrum to check middle peaks. Their phases should have been mostly corrected automatically.
- Press M to show nmrproc,com. Add the P0 and P1 values shown next to P0/P1 bars to the nmrproc.com script for that dimension. The first line with nmrPipe -fn-P0 XXX-P1 XXX is for X-axis and the second is for Y-axis. Add or subtract the P0 and P1phases to those already there. Click Save, Execute and Done. Press r to redraw contours to check phases again. Re-adjust P0 and P1 again if needed.
2: Procedure (snapshot during experiment with snap2d)
Nmrpipe Sol
Follow the same procedure above for data processing after experimentfinishes, except the following:
- Make a temporary folder under your home directory and go inside that folder. All temporary raw and processed data will be contained in this folder. AS an example, we name the folder as temp.fid.
- cd (change to home directory)
- mkdir temp.fid (create temp.fid folder)
- cd temp.fid (change directory to inside temp.fid)
- Then, type:
- snap2d
The program asks you to enter:
- Current vnmrJ experiment number being run. This number is displayed in the upper left corner of the spectrum window with name like exp1 or exp2, etc. Only enter the number after exp.
- Template experiment index number from the list (Default is 0 if you simply hit Enter/Return key)
- Current accumulated FID number. This number is updated at the bottom of the vnmrJ window as the experiment runs.
Once the information is provided, the program creates and runs the scriptsand launches nmrDraw. Press r with cursor in nmrDraw window todraw contours.
You can stop the experiment (with aa or click AbortAcquisition button) anytime if you feel the data quality is adequatefrom the snapshot. Note that if a 2D data collection aborts before the last FIDis collected, the data matrix is only filled partially. There appears to be nosaved parameter in the parameter file procpar that indicatesthe index number of the last FID collected. nmrPipe gives an error if the full2D matrix is to be processed. snap2D asks user input for thecorrect size of finished FIDs. But if you do not know this value after datacollection, you can reduce the Y-axis points (-yN and-yT fields) via try-and-error and re-run the data conversionuntil the error message disppears.
After you finish with the experiment, to make a copy of the snapshot datafolder as your final data folder, type:
- cd . (the two dots indicate move up one folder)
- cp -r temp.fid my_final_data.fid
Alternatively, you can rename the temp folder to the final data folder namerather than copy it to a new folder:
Nmrpipe Coadd
- mv temp.fid my_final_data.fid
3: Plot 2D spectrum with X and Y projections
To generate a plot with the X and Y projectiondisplayed along the edges, type the following commands:
- cd noesy2d.fid (change directory to inside the .fid folder)
- plot1D2D.tcl
- ps2pdf roi.ps
By default, the plot1D2D.tcl program takestest.ft2 as input, automatically set contour threshold andplot the whole region of the spectrum with projections along the sides. Thedefault output postscript file is: roi.ps. Theps2pdf command turns the .ps file into a pdf file(roi.pdf). Both can be edited with Adobe Illustrator or otherprograms without the loss of digital resolution.
To plot only a region of the spectrum or use a different contour threshold(find the value next to First: in nmrDraw display), modify thefollowing commands: Sun java runtime environment 1.7 0.
Nmrpipe Download
- plot1D2D.tcl -x1 2.0 -xn 5.0 -y1 2.0 -yn 5.0 (plot region from 2 to 5ppm along both X and Y)
- plot1D2D.tcl -hi 50000.0 -x1 2.0 -xn 5.0 -y1 3.0 -yn 8.0 (plot with a threshold of 50000, from 2 to 5ppm along X and 3 to 8ppm along Y)
- plot1D2D.tcl -out expand.ps-x1 2.0-xn 5.0 (plot from 2 to 5ppm along X and all along Y, with output file named as expand.ps)
Nmrpipe Sp
If any of the options starting with a dash ('-') is not specified, thedefault value or name is used. See the entire list of the options from Frank Delaglio'swebsite here.
Nmrpipe Download
4: Reference PPM adjustment
Nmrdraw
pipe2d and snap2d automatically setreference PPM for the indirect dimension according to IUPAC unified chemicalshift method. The method relies on the presence of accurate referenceparameters of direct 1H dimension saved (from vnmrJ) in the parameter fileprocpar. If for some reason, you forget to keep properreferencing before saving the data, use one of the several methods below tocorrect the reference PPM during processing.
- If you know the PPM value of a 1H signal in the spectrum
- The best way is to modify fid.com and re-do the data conversion. Open fid.com with a text editor (nedit fid.com&), and add or subtract the offset PPM value to -xCAR or -yCAR field to give the correct PPM value for the reference peak. Save the file. Then, type fid.com, followed by nmrproc.com. Check the correction in nmrDraw.
- Alternatively, from nmrDraw, right button click File->Calibrate Axis. Select the axis you want to adjust (either X or Y). Enter a new reference PPM value for that axis. You should add or subtract the offset between the your current peak PPM and the correct PPM value to the reference value field. Then, click Apply, and Save. Note that this method changes the reference parameters in the processed data (test.ft2) only, but not in the converted raw data (test.fid). Therefore, if you re-process the data with nmrproc.com, the reference correction is overwritten.
- If you do not know anything about the PPM value of the compound, the best is to re-collect a 1D 1H spectrum under the same condition, with TMS in your sample or less precisely use the lock (or residual solvent peak or simply enter setref) to get the reference right. It is possible to back calculate the PPM values based on the saved parameters (lock position), tof and dof offset etc., but the uncertainty could be substantial.
Nmrpipe Nmr
H. Zhouupdated Oct 2010